CV
Download full CVEducation
After graduating high school, I took a few personal years (a year of medical school, small jobs, and travelling alone with my backpack). I then followed my original passion and studied biology at Toulouse University. After a Bachelor in Biology of Organisms, Populations & Ecosystems (BOPE), that gave me an integrated view of biological mechanisms, I specialized in molecular biology with a MSc in Plants Adaptation, Development & Improvement in presence of Microorganisms (ADAM). In 2025, I obtained my PhD degree from the Department of Plant development, Biotic and Abiotic Interactions.

Research Overview
During my MSc, I studied the regulation of nodule organogenesis by a lncRNA located in within a symbiotic island and potentially targeted by NF-YA1, in the Medicago-Sinorhizobium symbiotic system, under the supervision of Dr. Andreas Niebel.
After my MSc, I chose to work on plant-pathogen interactions and secured a fellowship to develop my PhD project with Dr. Laurent Deslandes (Group leader, PhD co-supervisor) and Dr. Valérie Pacquit (Senior lecturer, PhD co-supervisor). My work aimed at uncovering the extent of bacterial effectors virulence strategies, with a particular focus on the homolog effectors PopP2 from Ralstonia pseudosolanacearum and XopJ6 from Xanthomonas campestris, both already known to target WRKY transcription factors in Arabidopsis thaliana.



-
Main results
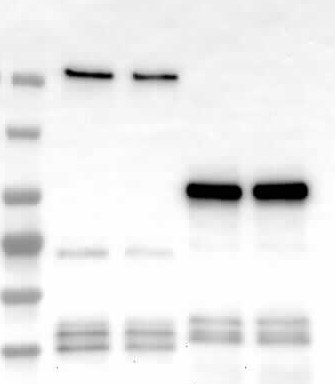
- We identified and characterized three ARID proteins as additional virulence targets of PopP2 (and possibly of XopJ6 as well) demonstrating that PopP2 can target diverse host proteins. Our findings also suggested a manipulation of chromatin remodelling complexes by PopP2 through ARIDs targeting —a virulence mechanism rarely described for effectors from phytopathogenic bacteria.
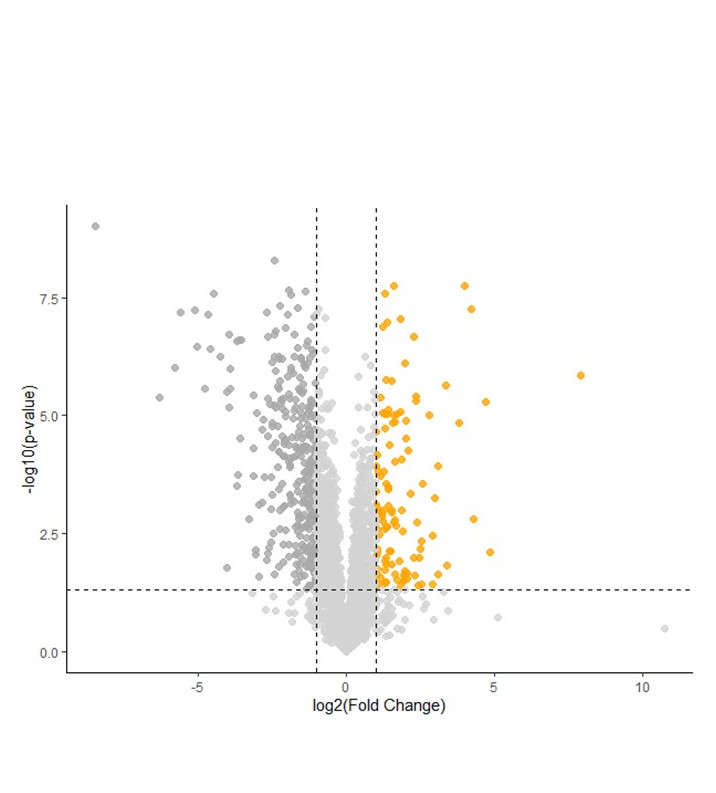
- I used proximity labelling to identify PopP2 and XopJ6 proximal proteomes in Arabidopsis thaliana. The proteomes revealed exciting additional potential targets, some of which are shared between the two effectors. A manuscript detailing our findings will be submitted shortly.
- Overall, we gained insights into the diverse host functions that could be subverted by these YopJ effectors from two distinct xylem-colonizing bacteria.
